Recurrent Models and Examples with MXNetR
As a new lightweight and flexible deep learning platform, MXNet provides a portable backend, which can be called from R side. MXNetR is an R package that provide R users with fast GPU computation and state-of-art deep learning models.
In this post, We have provided several high-level APIs for recurrent models with MXNetR. Recurrent neural network (RNN) is a class of artificial neural networks, which is very popular in the sequence labelling tasks, such as handwriting recognition, speech recognition.
We will introduce our implementation of the recurrent models including RNN, LSTM and GRU. In addition, the examples such as char-rnn will be showed to explain how to use the RNN models. By the way, several optimizers are added in MXNetR too.
This post demonstrates the implementation of the recurrent modules, including the structure of recurrent cells, unrolling of RNN models and the specific functions for training and testing.
-
Three kinds of recurrent cells, including custom RNN, LSTM and GRU cells.
-
How to unroll RNN models to common feedward network.
-
How to train RNN models, including setting up and training RNN models by our specific solver using low-level simple-bind interface.
-
How to utilize the trained RNN models using RNN inference interfaces, including
inferenceandforwardfunctions.
The link to the commits is https://github.com/dmlc/mxnet/commits?author=ziyeqinghan.
Recurrent Models
Since the RNN model can be treated as a deep feedforward neural network, which unfolds in time, it suffers from the problem of vanishing and exploding gradients. Thus, there are several variants of RNN to learn the long term dependency, including Long Short-Term Memory (LSTM) [1] and Gated Recurrent Unit (GRU) [2].
We will introduce three RNN models including the custom RNN, LSTM and GRU which has been implemented in MXNetR. To see the complete code, please refer to the relevant files rnn.R, lstm.R, gru.R and rnn_model.R in the R-package/R directory respectively.
RNN Cells
The main difference between three RNN models is that they have corresponding cells with different structures to mitigate the problem of vanishing and exploding gradients.
Custom RNN Cells
The Common RNN can be considered as feedforward networks with self-connected hidden layers. As Figure 1 shows, the key of the RNN is that it allows the previous inputs has influence on the current output by using the recurrent connection.
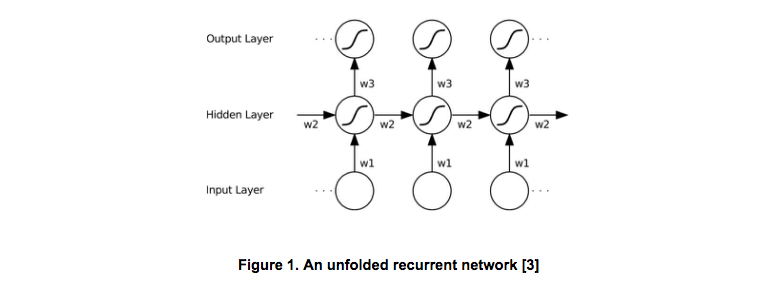
Given an input sequence x and the previous state h, a custom RNN cell produces the next states successively. Thus, there are two types of connections, the input to the hidden i2h, and the (previous) hidden to the hidden h2h. Then an optional batch normalization layer and nonlinear activation layer (e.g. tanh) are followed to generate the output states.
rnn <- function(num.hidden, indata, prev.state, param, seqidx,
layeridx, dropout=0., batch.norm=FALSE) {
if (dropout > 0. )
indata <- mx.symbol.Dropout(data=indata, p=dropout)
i2h <- mx.symbol.FullyConnected(data=indata,
weight=param$i2h.weight,
bias=param$i2h.bias,
num.hidden=num.hidden,
name=paste0("t", seqidx, ".l", layeridx, ".i2h"))
h2h <- mx.symbol.FullyConnected(data=prev.state$h,
weight=param$h2h.weight,
bias=param$h2h.bias,
num.hidden=num.hidden,
name=paste0("t", seqidx, ".l", layeridx, ".h2h"))
hidden <- i2h + h2h
hidden <- mx.symbol.Activation(data=hidden, act.type="tanh")
if (batch.norm)
hidden <- mx.symbol.BatchNorm(data=hidden)
return (list(h=hidden))
}
LSTM Cells
LSTM replace the cells in custom RNN with LSTM memory block. Figure 2 illustrates the architecture of an LSTM unit.
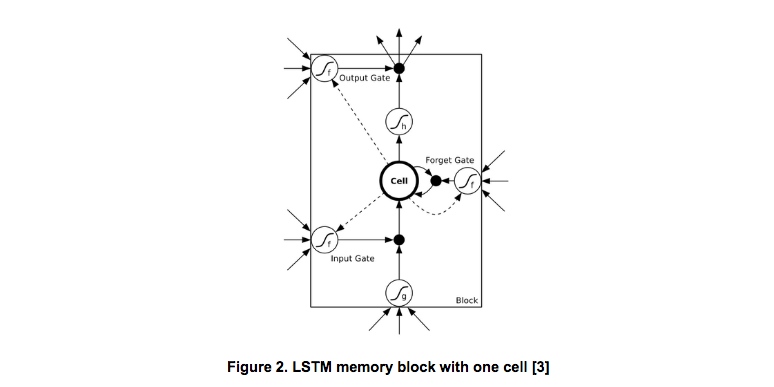
It contains one memory cell and three multiplicative units, i.e., the input gate, the forget gate and the output gate. With the help of the memory cell and the gates, LSTM can store and learn long term dependencies across the whole sequence.
A LSTM cell produces the next states by based on the input x as well as previous states (including c and h). For gates, there are three types of connections, including the input to the gate, the (previous) hidden to the gate and the cell to the gate. The activation functions of the gates should use the sigmoid function to make sure the outputs of gates in range [0, 1]. For the memory cell, there are two connections, the input to the cell and the (previous) hidden to the cell.
lstm <- function(num.hidden, indata, prev.state, param, seqidx, layeridx, dropout=0) {
if (dropout > 0)
indata <- mx.symbol.Dropout(data=indata, p=dropout)
i2h <- mx.symbol.FullyConnected(data=indata,
weight=param$i2h.weight,
bias=param$i2h.bias,
num.hidden=num.hidden * 4,
name=paste0("t", seqidx, ".l", layeridx, ".i2h"))
h2h <- mx.symbol.FullyConnected(data=prev.state$h,
weight=param$h2h.weight,
bias=param$h2h.bias,
num.hidden=num.hidden * 4,
name=paste0("t", seqidx, ".l", layeridx, ".h2h"))
gates <- i2h + h2h
slice.gates <- mx.symbol.SliceChannel(gates, num.outputs=4,
name=paste0("t", seqidx, ".l", layeridx, ".slice"))
in.gate <- mx.symbol.Activation(slice.gates[[1]], act.type="sigmoid")
in.transform <- mx.symbol.Activation(slice.gates[[2]], act.type="tanh")
forget.gate <- mx.symbol.Activation(slice.gates[[3]], act.type="sigmoid")
out.gate <- mx.symbol.Activation(slice.gates[[4]], act.type="sigmoid")
next.c <- (forget.gate * prev.state$c) + (in.gate * in.transform)
next.h <- out.gate * mx.symbol.Activation(next.c, act.type="tanh")
return (list(c=next.c, h=next.h))
}
Instead of defining the gates and the memory cell independently, we compute them together and then use mx.symbol.SliceChannel to split them into four outputs.
GRU Cells
GRU is another variant model of RNN, which was proposed in 2014 [2]. Similar to LSTM unit, the GRU unit also aims to adaptively capture dependencies of different time scales by updating and reseting gate as shown in Figure 3.
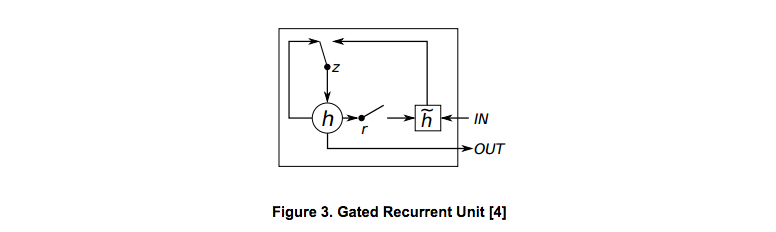
The calculation of GRU is similar with the custom RNN and LSTM models. First, there are two types of connections for gates to be defined: update.gate decides how much the unit updates its activations and reset.gate which decides whether to forget the previously computed state. Then, the candidate activation htrans is computed using input data x and previous state h (reset.gate controls whether to forget the previous state). After getting htrans, use update.gate to decide the proportion of htrans and previous state h to calculate next state h.
gru <- function(num.hidden, indata, prev.state, param, seqidx, layeridx, dropout=0) {
if (dropout > 0)
indata <- mx.symbol.Dropout(data=indata, p=dropout)
i2h <- mx.symbol.FullyConnected(data=indata,
weight=param$gates.i2h.weight,
bias=param$gates.i2h.bias,
num.hidden=num.hidden * 2,
name=paste0("t", seqidx, ".l", layeridx, ".gates.i2h"))
h2h <- mx.symbol.FullyConnected(data=prev.state$h,
weight=param$gates.h2h.weight,
bias=param$gates.h2h.bias,
num.hidden=num.hidden * 2,
name=paste0("t", seqidx, ".l", layeridx, ".gates.h2h"))
gates <- i2h + h2h
slice.gates <- mx.symbol.SliceChannel(gates, num.outputs=2,
name=paste0("t", seqidx, ".l", layeridx, ".slice"))
update.gate <- mx.symbol.Activation(slice.gates[[1]], act.type="sigmoid")
reset.gate <- mx.symbol.Activation(slice.gates[[2]], act.type="sigmoid")
htrans.i2h <- mx.symbol.FullyConnected(data=indata,
weight=param$trans.i2h.weight,
bias=param$trans.i2h.bias,
num.hidden=num.hidden,
name=paste0("t", seqidx, ".l", layeridx, ".trans.i2h"))
h.after.reset <- prev.state$h * reset.gate
htrans.h2h <- mx.symbol.FullyConnected(data=h.after.reset,
weight=param$trans.h2h.weight,
bias=param$trans.h2h.bias,
num.hidden=num.hidden,
name=paste0("t", seqidx, ".l", layeridx, ".trans.h2h"))
h.trans <- htrans.i2h + htrans.h2h
h.trans.active <- mx.symbol.Activation(h.trans, act.type="tanh")
next.h <- prev.state$h + update.gate * (h.trans.active - prev.state$h)
return (list(h=next.h))
}
Unrolling RNN Models
Since MXNet has implemented low level layers, we can unroll the RNN model in the time dimension and then use the MXNet layers to construct the different RNN networks according the above defined RNN cells. After unrolling the RNN in time, the model is just like the common feedforward network unless the weights shared in different feedforward layers and specific different recurrent units. Specifically, we can use fully-connected layers and corresponding activations to represent the different types of connections.
The unroll function needs to unroll the recurrent model according to the predefined sequence length. The recurrent weights across time need to be shared and the network depth represents the number of recurrent layers.
We provide the unrolling functions which is suitable for tasks like character language model and PennTreeBank language model. For other tasks the unrolling functions are similiar.
First, we define the weights and states. embed.weight is the weights used for embedding layer, mapping from the one-hot input to a dense vector. cls.weight and cls.bias are the weights and bias for the final prediction at each time step. param.cells and last.states is the weights and states for each cell. The weights are shared and the states are different and will be updates over time.
lstm.unroll <- function(num.lstm.layer, seq.len, input.size,
num.hidden, num.embed, num.label, dropout=0.) {
embed.weight <- mx.symbol.Variable("embed.weight")
cls.weight <- mx.symbol.Variable("cls.weight")
cls.bias <- mx.symbol.Variable("cls.bias")
param.cells <- lapply(1:num.lstm.layer, function(i) {
cell <- list(i2h.weight = mx.symbol.Variable(paste0("l", i, ".i2h.weight")),
i2h.bias = mx.symbol.Variable(paste0("l", i, ".i2h.bias")),
h2h.weight = mx.symbol.Variable(paste0("l", i, ".h2h.weight")),
h2h.bias = mx.symbol.Variable(paste0("l", i, ".h2h.bias")))
return (cell)
})
last.states <- lapply(1:num.lstm.layer, function(i) {
state <- list(c=mx.symbol.Variable(paste0("l", i, ".init.c")),
h=mx.symbol.Variable(paste0("l", i, ".init.h")))
return (state)
})
Then we unroll the RNN model in the time dimension and then use the MXNet layers to construct the RNN networks. Here the mx.symbol.Embedding is used to get the embedding vector for the specific task (char-rnn) shown there. At each time step, we share the weights param.cells and update the states last.states. Also, last.hidden is used to collect the outputs over time.
# embeding layer
label <- mx.symbol.Variable("label")
data <- mx.symbol.Variable("data")
embed <- mx.symbol.Embedding(data=data, input_dim=input.size,
weight=embed.weight, output_dim=num.embed, name="embed")
wordvec <- mx.symbol.SliceChannel(data=embed, num_outputs=seq.len, squeeze_axis=1)
last.hidden <- list()
for (seqidx in 1:seq.len) {
hidden <- wordvec[[seqidx]]
# stack lstm
for (i in 1:num.lstm.layer) {
dp <- ifelse(i==1, 0, dropout)
next.state <- lstm(num.hidden, indata=hidden,
prev.state=last.states[[i]],
param=param.cells[[i]],
seqidx=seqidx, layeridx=i,
dropout=dp)
hidden <- next.state$h
last.states[[i]] <- next.state
}
# decoder
if (dropout > 0)
hidden <- mx.symbol.Dropout(data=hidden, p=dropout)
last.hidden <- c(last.hidden, hidden)
}
Finally, we need to construct the remain layers according to the different tasks. Take char-rnn as an example, cls.weight and cls.bias are used for the final prediction and then mx.symbol.SoftmaxOutput connnets the prediction and corresponding labels to back propagate though time.
last.hidden$dim <- 0
last.hidden$num.args <- seq.len
concat <-mxnet:::mx.varg.symbol.Concat(last.hidden)
fc <- mx.symbol.FullyConnected(data=concat,
weight=cls.weight,
bias=cls.bias,
num.hidden=num.label)
label <- mx.symbol.transpose(data=label)
label <- mx.symbol.Reshape(data=label, target.shape=c(0))
loss.all <- mx.symbol.SoftmaxOutput(data=fc, label=label, name="sm")
return (loss.all)
}
Training RNN Models
After implementing unrolled RNN models, we need to know how to train the recurrent models. We write the training code on our own using the low level symbol interfaces instead of the common FeedForward model. The reason why we implement by ourselves is two-fold. First, currently, existing data iterators do not support sequence data well. Second, since our input contains both the input data x and the states i.e h and we need set the gradients of them to zero at each epoch, the existing high level FeedForward interface is not appropriate for our tasks.
For the reasons above, we write the training code on our own to bind the network and train epoch by epoch using the low level symbol interfaces. You can refer to the code in rnn_model.R. The training method of three RNN models are the same except the initial states are init.h or init.c and init.h. So they can use the same training function.
The training codes mainly contain the function setup.rnn.model and the function train.rnn.
Set Up RNN Models
setup.rnn.model is defined to initialize parameters and bind the network. The parameter init.states.name stores the name of the initial state. For custom RNN and GRU, init.states.name is end with init.h. For LSTM, init.states.name is end with init.h and init.c.
First, we set the dimension of the input including input data, label and initial states. Given the input dimension, we can use mx.model.init.params.rnn function to initialize parameters.
setup.rnn.model <- function(rnn.sym, ctx,
num.rnn.layer, seq.len,
num.hidden, num.embed, num.label,
batch.size, input.size,
init.states.name,
initializer=mx.init.uniform(0.01),
dropout=0) {
arg.names <- rnn.sym$arguments
input.shapes <- list()
for (name in arg.names) {
if (name %in% init.states.name) {
input.shapes[[name]] <- c(num.hidden, batch.size)
}
else if (grepl('data$', name) || grepl('label$', name) ) {
if (seq.len == 1) {
input.shapes[[name]] <- c(batch.size)
} else {
input.shapes[[name]] <- c(seq.len, batch.size)
}
}
}
params <- mx.model.init.params.rnn(rnn.sym, input.shapes, initializer, mx.cpu())
Next, we use mx.simple.bind to bind the network, set the arg.arrays, aux.arrays and grad.arrays.
args <- input.shapes
args$symbol <- rnn.sym
args$ctx <- ctx
args$grad.req <- "add"
rnn.exec <- do.call(mx.simple.bind, args)
mx.exec.update.arg.arrays(rnn.exec, params$arg.params, match.name=TRUE)
mx.exec.update.aux.arrays(rnn.exec, params$aux.params, match.name=TRUE)
grad.arrays <- list()
for (name in names(rnn.exec$ref.grad.arrays)) {
if (is.param.name(name))
grad.arrays[[name]] <- rnn.exec$ref.arg.arrays[[name]]*0
}
mx.exec.update.grad.arrays(rnn.exec, grad.arrays, match.name=TRUE)
return (list(rnn.exec=rnn.exec, symbol=rnn.sym,
num.rnn.layer=num.rnn.layer, num.hidden=num.hidden,
seq.len=seq.len, batch.size=batch.size,
num.embed=num.embed))
}
Training RNN Models
train.rnn function is to train network epoch by epoch, update the parameters using mx.exec.forward and mx.exec.backward, which is similar to the function mx.model.train in model.R. One difference is that in train.rnn, at the beginning of each epoch, the initial states should be clear.
for (name in init.states.name) {
init.states[[name]] <- m$rnn.exec$ref.arg.arrays[[name]]*0
}
mx.exec.update.arg.arrays(m$rnn.exec, init.states, match.name=TRUE)
Also, at the end of the epoch, when updating the gradient of parameters, the gradient of initial c and initial h should be zero.
# the gradient of initial c and inital h should be zero
init.grad <- list()
for (name in init.states.name) {
init.grad[[name]] <- m$rnn.exec$ref.arg.arrays[[name]]*0
}
mx.exec.update.grad.arrays(m$rnn.exec, init.grad, match.name=TRUE)
APIs of custom RNN, LSTM, GRU
mx.rnn, mx.lstm, mx.gru is the APIs for training custom RNN / LSTM / GRU unrolled model. They will first change the data into iterator and then get the unrolled rnn symbol. After getting the unrolled rnn symbol, the function sets up the training model, binds the network and then trains the model to get the final result.
We show the mx.lstm function, the other two are similar to it.
mx.lstm <- function(train.data, eval.data=NULL,
num.lstm.layer, seq.len,
num.hidden, num.embed, num.label,
batch.size, input.size,
ctx=mx.ctx.default(),
num.round=10, update.period=1,
initializer=mx.init.uniform(0.01),
dropout=0, optimizer='sgd',
...) {
# check data and change data into iterator
train.data <- check.data(train.data, batch.size, TRUE)
eval.data <- check.data(eval.data, batch.size, FALSE)
# get unrolled lstm symbol
rnn.sym <- lstm.unroll(num.lstm.layer=num.lstm.layer,
num.hidden=num.hidden,
seq.len=seq.len,
input.size=input.size,
num.embed=num.embed,
num.label=num.label,
dropout=dropout)
init.states.c <- lapply(1:num.lstm.layer, function(i) {
state.c <- paste0("l", i, ".init.c")
return (state.c)
})
init.states.h <- lapply(1:num.lstm.layer, function(i) {
state.h <- paste0("l", i, ".init.h")
return (state.h)
})
init.states.name <- c(init.states.c, init.states.h)
# set up lstm model
model <- setup.rnn.model(rnn.sym=rnn.sym,
ctx=ctx,
num.rnn.layer=num.lstm.layer,
seq.len=seq.len,
num.hidden=num.hidden,
num.embed=num.embed,
num.label=num.label,
batch.size=batch.size,
input.size=input.size,
init.states.name=init.states.name,
initializer=initializer,
dropout=dropout)
# train lstm model
model <- train.rnn( model, train.data, eval.data,
num.round=num.round,
update.period=update.period,
ctx=ctx,
init.states.name=init.states.name,
...)
# change model into MXFeedForwardModel
model <- list(symbol=model$symbol, arg.params=model$rnn.exec$ref.arg.arrays, aux.params=model$rnn.exec$ref.aux.arrays)
return(structure(model, class="MXFeedForwardModel"))
}
RNN Inference Models
In the tasks like character language model and PennTreeBank language model, we unroll rnn models with specific seq.len when training models. After we get the trained models, we may want to use it with different seq.len from the unrolled rnn models. For example, as for char-rnn model, we first train a char level language model with seq.len=128. Then we may want to generate a 100-length text from it. The weights of rnn cells are shared at each time step, so we only need to store a network with seq.len=1 after training.
RNN Inference APIs
mx.rnn.inference, mx.lstm.inference, mx.gru.inference is used to create a custom RNN / LSTM / GRU inference model with parameter seq.len=1. We show the codes of mx.lstm.inference and the other two are similar to it. The parameter arg.params stores the parameters of the trained unrolled rnn model. We build the network with seq.len=1. Then we set up the rnn model with initialization and binding. We use mx.exec.update.arg.arrays to set the arg.arrays to arg.params of the trained model. The initial states are cleared as well.
mx.lstm.inference <- function(num.lstm.layer,
input.size,
num.hidden,
num.embed,
num.label,
batch.size=1,
arg.params,
ctx=mx.cpu(),
dropout=0.) {
sym <- lstm.inference.symbol(num.lstm.layer=num.lstm.layer,
input.size=input.size,
num.hidden=num.hidden,
num.embed=num.embed,
num.label=num.label,
dropout=dropout)
init.states.c <- lapply(1:num.lstm.layer, function(i) {
state.c <- paste0("l", i, ".init.c")
return (state.c)
})
init.states.h <- lapply(1:num.lstm.layer, function(i) {
state.h <- paste0("l", i, ".init.h")
return (state.h)
})
init.states.name <- c(init.states.c, init.states.h)
seq.len <- 1
# set up lstm model
model <- setup.rnn.model(rnn.sym=sym,
ctx=ctx,
num.rnn.layer=num.lstm.layer,
seq.len=seq.len,
num.hidden=num.hidden,
num.embed=num.embed,
num.label=num.label,
batch.size=batch.size,
input.size=input.size,
init.states.name=init.states.name,
initializer=mx.init.uniform(0.01),
dropout=dropout)
arg.names <- names(model$rnn.exec$ref.arg.arrays)
for (k in names(arg.params)) {
if ((k %in% arg.names) && is.param.name(k) ) {
rnn.input <- list()
rnn.input[[k]] <- arg.params[[k]]
mx.exec.update.arg.arrays(model$rnn.exec, rnn.input, match.name=TRUE)
}
}
init.states <- list()
for (i in 1:num.lstm.layer) {
init.states[[paste0("l", i, ".init.c")]] <- model$rnn.exec$ref.arg.arrays[[paste0("l", i, ".init.c")]]*0
init.states[[paste0("l", i, ".init.h")]] <- model$rnn.exec$ref.arg.arrays[[paste0("l", i, ".init.h")]]*0
}
mx.exec.update.arg.arrays(model$rnn.exec, init.states, match.name=TRUE)
return (model)
}
RNN Forward APIs
After getting the inference model with seq.len=1, we use the forward functions (mx.rnn.forward, mx.lstm.forward, mx.gru.forward) to perform prediction in custom RNN / LSTM / GRU inference model. The parameter new.seq denotes that whether there is a new sequence and the previous states should be clear. Given the previous states and input data, we use mx.exec.forward to get the output. After forwarding, the states c and h should be stored to be the previous states for the next input.
mx.lstm.forward <- function(model, input.data, new.seq=FALSE) {
if (new.seq == TRUE) {
init.states <- list()
for (i in 1:model$num.rnn.layer) {
init.states[[paste0("l", i, ".init.c")]] <- model$rnn.exec$ref.arg.arrays[[paste0("l", i, ".init.c")]]*0
init.states[[paste0("l", i, ".init.h")]] <- model$rnn.exec$ref.arg.arrays[[paste0("l", i, ".init.h")]]*0
}
mx.exec.update.arg.arrays(model$rnn.exec, init.states, match.name=TRUE)
}
dim(input.data) <- c(model$batch.size)
data <- list(data=mx.nd.array(input.data))
mx.exec.update.arg.arrays(model$rnn.exec, data, match.name=TRUE)
mx.exec.forward(model$rnn.exec, is.train=FALSE)
init.states <- list()
for (i in 1:model$num.rnn.layer) {
init.states[[paste0("l", i, ".init.c")]] <- model$rnn.exec$ref.outputs[[paste0("l", i, ".last.c_output")]]
init.states[[paste0("l", i, ".init.h")]] <- model$rnn.exec$ref.outputs[[paste0("l", i, ".last.h_output")]]
}
mx.exec.update.arg.arrays(model$rnn.exec, init.states, match.name=TRUE)
prob <- model$rnn.exec$ref.outputs[["sm_output"]]
return (list(prob=prob, model=model))
}
RNN Example
All in all, we can use the APIs mx.lstm, mx.lstm.inference, mx.lstm.forward to train LSTM models, create a LSTM inference model and use forward function to predict in LSTM inference model. We provide a char-LSTM example. This example aims to show how to use LSTM model to build a char level language model, and generate text from it. We use a tiny shakespeare text for demo purpose. more details can refer to char-RNN documentation.
Load data
First of all, we load in the data and preprocess it. We transfer the test into data feature matrix. The dimension of the matrix is seq.len * num.seq, the element of the matrix is the id of the char.
We can get X.train ans X.val which represent the training data and evaluation data.
X.train <- list(data=X.train.data, label=X.train.label)
X.val <- list(data=X.val.data, label=X.val.label)
Training Model
In mxnet, we have a function called mx.lstm so that users can build a general lstm model.
Set basic network parameters.
batch.size = 32
seq.len = 32
num.hidden = 256
num.embed = 256
num.lstm.layer = 2
num.round = 5
learning.rate= 0.1
wd=0.00001
clip_gradient=1
update.period = 1
Training the LSTM model.
model <- mx.lstm(X.train, X.val,
ctx=mx.cpu(),
num.round=num.round,
update.period=update.period,
num.lstm.layer=num.lstm.layer,
seq.len=seq.len,
num.hidden=num.hidden,
num.embed=num.embed,
num.label=vocab,
batch.size=batch.size,
input.size=vocab,
initializer=mx.init.uniform(0.1),
learning.rate=learning.rate,
wd=wd,
clip_gradient=clip_gradient)
## Epoch [31] Train: NLL=3.47213018872144, Perp=32.2052727363657
## ...
## Epoch [961] Train: NLL=2.32060007657895, Perp=10.181782322355
## Iter [1] Train: Time: 161.368175268173 sec, NLL=2.31135356537961, Perp=10.0880702804858
## Iter [1] Val: NLL=1.94184484060012, Perp=6.97160060607419
## Epoch [992] Train: NLL=1.84784553299322, Perp=6.34613225095329
## ...
## Epoch [1953] Train: NLL=1.70175791172558, Perp=5.48357857093351
## Iter [2] Train: Time: 163.582908391953 sec, NLL=1.70103940328978, Perp=5.47963998859367
## Iter [2] Val: NLL=1.74979316010449, Perp=5.75341251767988
## Epoch [1984] Train: NLL=1.59619252884271, Perp=4.9342097524275
## ...
## Epoch [2914] Train: NLL=1.54738185300295, Perp=4.69915099483974
## Iter [3] Train: Time: 163.555737018585 sec, NLL=1.54604189517013, Perp=4.69285854740519
## Iter [3] Val: NLL=1.67780240235925, Perp=5.35377758479576
## Epoch [2945] Train: NLL=1.48868466087876, Perp=4.43126307034767
## ...
## Epoch [3906] Train: NLL=1.474871752958, Perp=4.37047523646576
## Iter [4] Train: Time: 155.97230219841 sec, NLL=1.4744973925858, Perp=4.36883940994296
## Iter [4] Val: NLL=1.64488167325603, Perp=5.18039689118454
## Epoch [3937] Train: NLL=1.46355541021581, Perp=4.32129622881604
## ...
## Epoch [4898] Train: NLL=1.42900458455642, Perp=4.17454171976281
## Iter [5] Train: Time: 151.142980337143 sec, NLL=1.42909226256273, Perp=4.17490775130428
## Iter [5] Val: NLL=1.62716655804022, Perp=5.08943365437187
Predict from the Pretrained Model
We define the choice function to get random samples. We can use random output or fixed output by choosing largest probability.
make.output <- function(prob, sample=FALSE) {
if (!sample) {
idx <- which.max(as.array(prob))
}
else {
idx <- choice(prob)
}
return (idx)
}
In mxnet, we have a function called mx.lstm.inference so that users can build a inference from lstm model and then use function mx.lstm.forward to get forward output from the inference.
Build inference from model.
infer.model <- mx.lstm.inference(num.lstm.layer=num.lstm.layer,
input.size=vocab,
num.hidden=num.hidden,
num.embed=num.embed,
num.label=vocab,
arg.params=model$arg.params,
ctx=mx.cpu())
Generate a sequence of 75 chars using function mx.lstm.forward.
start <- 'a'
seq.len <- 75
random.sample <- TRUE
last.id <- dic[[start]]
out <- "a"
for (i in (1:(seq.len-1))) {
input <- c(last.id-1)
ret <- mx.lstm.forward(infer.model, input, FALSE)
infer.model <- ret$model
prob <- ret$prob
last.id <- make.output(prob, random.sample)
out <- paste0(out, lookup.table[[last.id]])
}
cat (paste0(out, "\n"))
The result:
ah not a drobl greens
Settled asing lately sistering sounted to their hight
Other things new
optimizer.R in the R package is used to control the learning rate and update the weights during the training period. At past, only the sgd (Stochastic Gradient Descent) function was implemented. Right now, several other useful optimizer functions are added. The new added optimizers are:
- RMSProp optimizer
- Adam optimizer
- AdaGrad optimizer
- AdaDelta optimizer
Reference
[1] Hochreiter, Sepp, and Jürgen Schmidhuber. “Long short-term memory.”Neural computation 9.8 (1997): 1735-1780.
[2] Cho, Kyunghyun, et al. “On the properties of neural machine translation: Encoder-decoder approaches.” arXiv preprint arXiv:1409.1259 (2014).
[3] Graves, Alex. Supervised sequence labelling. Springer Berlin Heidelberg, 2012
[4] Chung, Junyoung, et al. “Empirical evaluation of gated recurrent neural networks on sequence modeling.” arXiv preprint arXiv:1412.3555 (2014).
